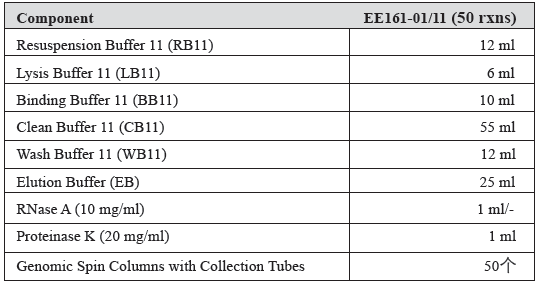
EasyPure® Bacteria Genomic DNA Kit (With RNase A)
Referencia EE161-01
embalaje : 50rxns
Marca : TransGen Biotech
EasyPure® Bacteria Genomic DNA Kit (with RNase A)
Catalog Number: EE161-01
Price:Please inquire first
Product Details
EasyPure® Bacteria Genomic DNA Kit uses lysozyme and moderate lysis buffer to lyse cells. Proteinase K is used for protein digestion and RNase A used for RNA digestion. DNA is specifically bound to silica-based column in hypersaline condition, and DNA is eluted by low salt and high pH solution. This kit is suitable for isolating high quality genomic DNA from Gram-positive and Gram-negative bacteria. The isolated DNA is suitable for PCR, restriction enzyme digestion, and Southern blot.
• Fast: the whole process can be completed in 50 minutes
• High yield: DNA yield up to 20 μg
Storage
At room temperature (15-25°C) in a dry place for one year.
Shipping
At room temperature
Liu Y, Chen Z, Jiang Z, et al. Biochemical characterization of a novel L-Asparaginase from Paenibacillus barengoltzii being suitable for acrylamide reduction in potato chips and mooncakes[J]. International Journal of Biological Macromolecules,2017, 104(Pt A):1055-1063.
Shi R , Liu Y , Mu Q , et al. Biochemical characterization of a novel L-asparaginase from Paenibacillus barengoltzii being suitable for acrylamide reduction in potato chips and mooncakes[J]. International Journal of Biological Macromolecules, 2017, 96:93-99.
Zheng X , Liu H , Song L , et al. Phaeocystidibacter marisrubri sp. nov. a member of the family Cryomorphaceae isolated from Red Sea sediment[J]. International Journal of Systematic and Evolutionary Microbiology, 2015, 65(7):2199.
Duan D , Cheng T . Determination of the microbial community features of Haemaphysalis flava in different developmental stages by high‐throughput sequencing[J]. Journal of Basic Microbiology, 2016, 57(4):302.
Cheng T Y , Liu G H . PCR denaturing gradient gel electrophoresis as a useful method to identify of intestinal bacteria flora in Haemaphysalis flava ticks[J]. Acta Parasitologica, 2017, 62(2):269-272.