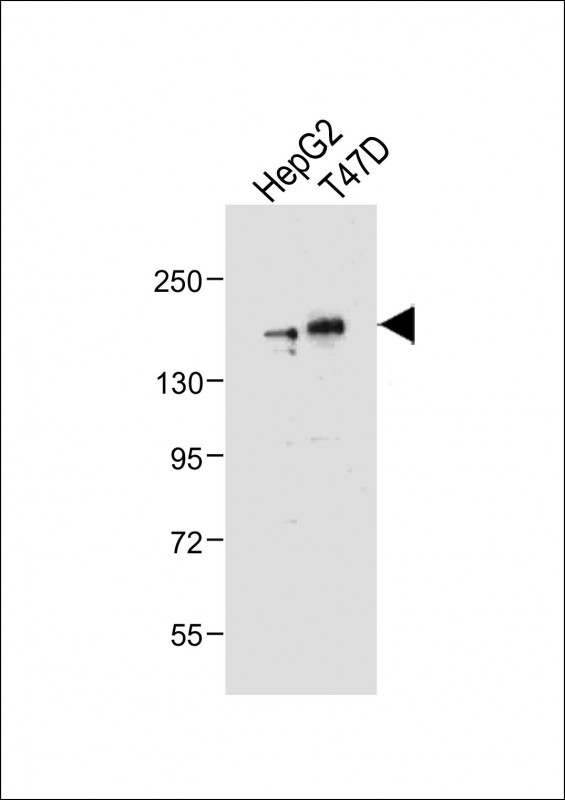
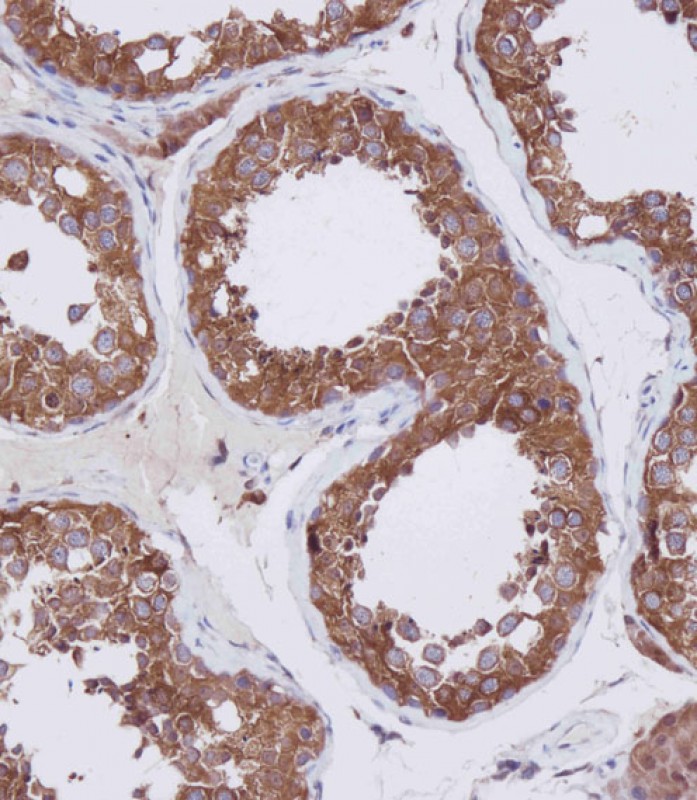
Application 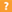
| IHC-P, WB, E |
|---|---|
| Primary Accession | Q9UBN7 |
| Other Accession | NP_006035 |
| Reactivity | Human |
| Host | Rabbit |
| Clonality | Polyclonal |
| Isotype | Rabbit IgG |
| Calculated MW | 131419 Da |
| Antigen Region | 1182-1215 aa |
| Gene ID | 10013 |
|---|---|
| Other Names | Histone deacetylase 6, HD6, HDAC6, KIAA0901 |
| Target/Specificity | This HDAC6 antibody is generated from rabbits immunized with a KLH conjugated synthetic peptide between 1182-1215 amino acids from the C-terminal region of human HDAC6. |
| Dilution | WB~~1:2000 IHC-P~~1:100 |
| Format | Purified polyclonal antibody supplied in PBS with 0.09% (W/V) sodium azide. This antibody is purified through a protein A column, followed by peptide affinity purification. |
| Storage | Maintain refrigerated at 2-8°C for up to 2 weeks. For long term storage store at -20°C in small aliquots to prevent freeze-thaw cycles. |
| Precautions | HDAC6 Antibody (C-term) is for research use only and not for use in diagnostic or therapeutic procedures. |
| Name | HDAC6 {ECO:0000303|PubMed:10220385, ECO:0000312|HGNC:HGNC:14064} |
|---|---|
| Function | Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4) (PubMed:10220385). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events (PubMed:10220385). Histone deacetylases act via the formation of large multiprotein complexes (PubMed:10220385). In addition to histones, deacetylates other proteins, such as CTTN, tubulin and SQSTM1 (PubMed:12024216, PubMed:20308065, PubMed:26246421, PubMed:30538141, PubMed:31857589). Plays a central role in microtubule-dependent cell motility by mediating deacetylation of tubulin (PubMed:12024216, PubMed:20308065, PubMed:26246421). Required for cilia disassembly; via deacetylation of alpha-tubulin (PubMed:17604723, PubMed:26246421). Promotes deacetylation of CTTN, leading to actin polymerization, promotion of autophagosome-lysosome fusion and completion of autophagy (PubMed:30538141). Involved in the MTA1-mediated epigenetic regulation of ESR1 expression in breast cancer (PubMed:24413532). Promotes odontoblast differentiation following IPO7-mediated nuclear import and subsequent repression of RUNX2 expression (By similarity). In addition to its protein deacetylase activity, plays a key role in the degradation of misfolded proteins: when misfolded proteins are too abundant to be degraded by the chaperone refolding system and the ubiquitin-proteasome, mediates the transport of misfolded proteins to a cytoplasmic juxtanuclear structure called aggresome (PubMed:17846173). Probably acts as an adapter that recognizes polyubiquitinated misfolded proteins and target them to the aggresome, facilitating their clearance by autophagy (PubMed:17846173). |
| Cellular Location | Cytoplasm. Cytoplasm, cytoskeleton. Nucleus {ECO:0000250|UniProtKB:Q9Z2V5}. Perikaryon {ECO:0000250|UniProtKB:Q9Z2V5}. Cell projection, dendrite {ECO:0000250|UniProtKB:Q9Z2V5}. Cell projection, axon {ECO:0000250|UniProtKB:Q9Z2V5}. Cell projection, cilium. Cytoplasm, cytoskeleton, microtubule organizing center, centrosome. Cytoplasm, cytoskeleton, cilium basal body. Note=It is mainly cytoplasmic, where it is associated with microtubules |

Provided below are standard protocols that you may find useful for product applications.
Background
HDAC6 (histone deacetylase 6) is responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Histone deacetylases act via the formation of large multiprotein complexes. HDAC6 plays a central role in microtubule-dependent cell motility via deacetylation of tubulin, and has been shown to interact with HDAC11, SIRT2, and F-actin. HDAC6 is ubiquitinated, but its polyubiquitination however does not lead to degradation. HDAC is also a potential target of sumoylation.
References
Hook, S.S., et al., Proc. Natl. Acad. Sci. U.S.A. 99(21):13425-13430 (2002).
Grozinger, C.M., et al., Proc. Natl. Acad. Sci. U.S.A. 96(9):4868-4873 (1999).
Wolffe, A.P., Nature 387(6628):16-17 (1997).
Pazin, M.J., et al., Cell 89(3):325-328 (1997).
Mahlknecht, U., et al., Cytogenet. Cell Genet. 93 (1-2), 135-136 (2001).