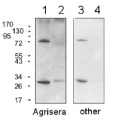
Anti-H+ATPase - Plasma membrane H+ATPase (rabbit antibody)
Referência AS07260
Tamanho : 50ug
Marca : Agrisera
1
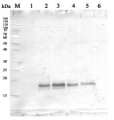
Anti-H+ATPase | Plasma membrane H+ATPase (rabbit antibody)
AS07 260 | Clonality: Polyclonal | Host: Rabbit | Reactivity: [global antibody] for di- and monocots, conifers, ferns, mosses, green algae | Cellular [compartment marker] for plasma membrane
Benefits of using this antibody
- Product Info
-
Immunogen: KLH-conjugated synthetic peptide, derived from available di and monocot, fern, mosses and algal plasma membrane ATPase sequences including Arabidopsis thaliana ATPase 1 (UniProt: P20649, TAIR: At2g18960) and ATPase 2 (UniProt: P19456 , TAIR: At4g30190), 3 (UniProt: P20431, TAIR: At5g57350), 4 (UniProt: Q9SU58, TAIR: At3g47950), 6 (UniProt: Q9SH76, TAIR: At2g07560), 7 (UniProt: Q9LY32, TAIR: At3g60330), 8 (UniProt: Q9M2A0, TAIR: At3g42640), 9 (UniProt: Q42556, TAIR: At1g80660), 11 (UniProt: Q9LV11, TAIR: At5g62670) of Arabidopsis thaliana and hydrogen ATPase of Chlamydomonas reinhardtii (Q9FNS3)
Host: Rabbit Clonality: Polyclonal Purity: Immunogen affinity purified serum in PBS pH 7.4. Format: Lyophilized Quantity: 50 µg Reconstitution: For reconstitution add 50 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Do not Store this antibody in 4°C. Tested applications: Immunofluorescence (IF), Immunolocalization (IL), Western blot (WB) Recommended dilution: 1 : 600-1 : 1000 (IF), 1 : 100 (IL), 1 : 1000-1 : 10 000 (WB) Expected | apparent MW: 90-95 kDa (Arabidopsis thaliana, depending upon an isoform)
- Reactivity
-
Confirmed reactivity: Actinidia chinensis, Aesculus hippocastanum, Arabidopsis thaliana, Camellia sinensis cv. Shu-chazao, Chara australis R.Br, Chlamydmonas reinhardtii, Cucumis sativus, Cucurbita moschata, Glycine max, Kandelia obovata, Hordeum vulgare, Lolium perenne, Lycopersicon esculentum, Malus x domestica Borkh. c.v. Fuji, Marchantia polymorpha, Medicago truncatula, Nicotiana benthamiana, Nicotiana tabacum, Noccaea caerulescens, Oryza sativa, Petunia hybrida, Phalenopsis Sogo Yukidian cultivar V3, Physcomitrium patens, Picea abies, Pisum sativum, Populus tremula, Pteris vittata (fern), Ricinus communis, Salicornia bigelovii, Spinacia oleracea, Solanum lycopersicum, Tagetes erecta, Tetraselmis chuii, Vicia faba, Zea mays Predicted reactivity: Amaranthus hypochondriacus,Avena sativa,Beta vulgaris, Cyanidioschyzon merolae, Dunaliella spp., Galdieria sulphuraria, Gossypium hirsutum, Hordeum vulgare, Ostreococcus spp., Pinus thunbergii, Physocomitrella patens, Mesembruanthemum crystallinum, Mortierella elongata, Nannochloropsis gaditana CCMP526, Ostreococcus tauri, Prosopis alba, Rosa chinensis Jacq., Saccharomyces cerevisiae, Solanum tuberosum, Sorghum bicolor, Spinacia oleracea, Triticum aestivum, Ulva prolifera, Ustilago maydis
Species of your interest not listed? Contact usNot reactive in: Allium sp., Aspergillus niger, Citrus limon, Colobanthus apetala, Cuminium cyminum, Curcuma amada, Deschampsia antractica, Lupinus luteus, Morinda citrifolia, Trigonella foenum, Vicia faba
- Application Examples
-
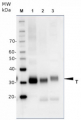
20 µg of total protein from Arabidopsis thaliana (1), Hordeum vulgare (2), Zea mays (3), Nicotiana tabaccum plasma membrane fraction, 2.5 µg (4), extracted with Protein Extration Buffer, PEB (AS08 300, homogenate the tissue with 3 to 5 volumes of the homogenizing buffer), were denaturated for 10 min. in 70°C and separated on 4-12% NuPage (Invitrogen) LDS-PAGE and blotted 1h to PVDF. Blots were blocked immediately following transfer in blocking reagent in 20 mM Tris, 137 mM sodium chloride pH 7.6 with 0.1% (v/v) Tween-20 (TBS-T) for 1h at room temperature with agitation. Blots were incubated in the primary antibody at a dilution of 1: 5 000 for 1h at room temperature with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, then washed once for 15 min and 3 times for 5 min in TBS-T at room temperature with agitation. Blots were incubated in secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated, recommended secondary antibody AS09 602) diluted to 1:20 000 in 2% blocking solution for 1h at room temperature with agitation. The blots were washed as above and developed for 5 min with chemiluminescence detection reagent according the manufacturers instructions. Images of the blots were obtained using a CCD imager (FluorSMax, Bio-Rad) and Quantity One software (Bio-Rad). Exposure time was 2 min.
Immunolocalization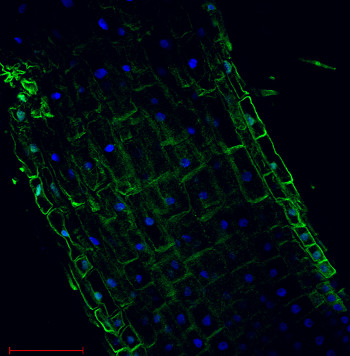
Plasma membrane H+ATPase localization inArabidopsis thaliana roots.
Arabidopsis thaliana, elongation zone, H+ATPase (green). Arabidopsis thaliana roots were fixed in para-formaldehyde for 30 minutes. Tissue cleaning has been performed before immunolocalization. Anti-rabbit H+ATPase | plasma membrane primary antibody diluted in 1: 300 and anti-rabbit IgG secondary antibody conjugated with Alexa 555. Co-staining with DAPI visualized nucleus (blue color). Scale bar – 100 µm.Courtesy Dr. Taras Pasternak, Freiburg University, Germany
Application examples: 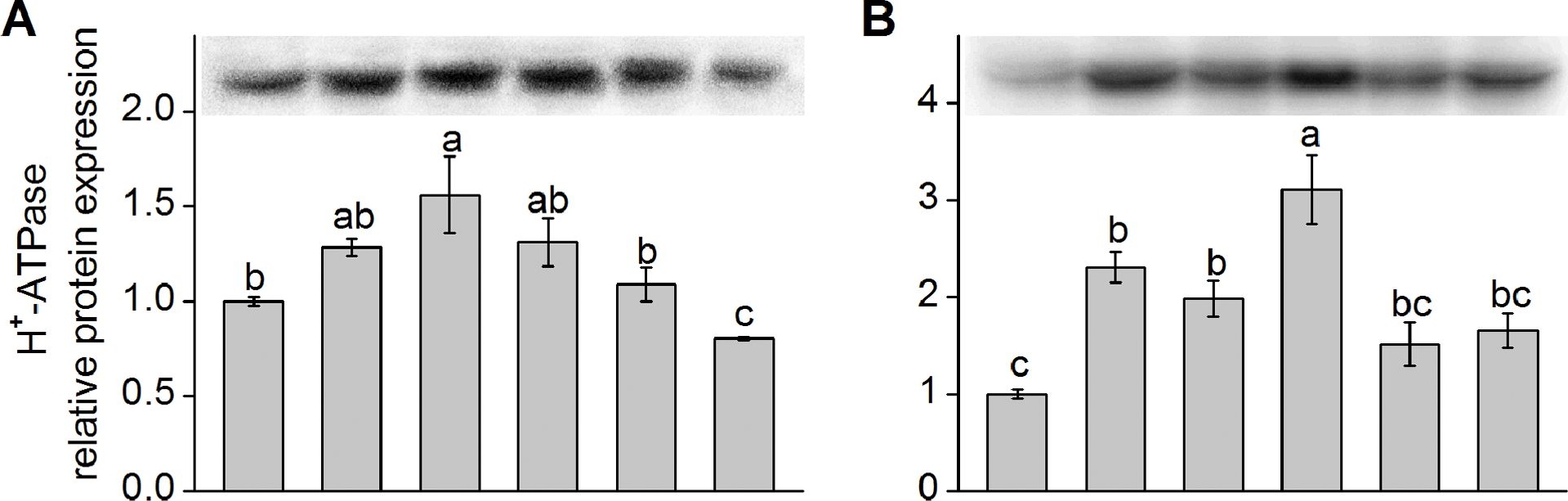
Reactant: Kandelia obovata
Application: Western Blotting
Pudmed ID: 23977070
Journal: PLoS One
Figure Number: 4A,B
Published Date: 2013-08-27
First Author: Chen, J., Xiong, D. Y., et al.
Impact Factor: 2.942
Open PublicationWestern blot analysis of protein expression of plasma membrane (PM) H+-ATPase and Na+/H+ antiporter (NHE-1).Shown in (A) and (C) are the relative protein expression level of K. obovata seedlings grown under 400 mM NaCl and various concentrations of SNP for 15 days. Shown in (B) and (D) represent the relative protein expression level of K. obovata seedlings treated with the following Hoagland solution-based solutions for 15 days: Control (only Hoagland solution), NaCl (400 mM), SNP (100 µM), NaCl+SNP (400 mM NaCl+100 µM SNP), cPTIO+L-NNA (200 µM cPTIO+100 µM L-NNA), NaCl+cPTIO+L-NNA (400 mM NaCl+200 µM cPTIO+100 µM L-NNA). Three independent experiments were performed. The relative intensity of bands (column charts) was quantified by densitometric analysis with Quantity One software and expressed relative to the intensity of the control band defined as 1. For each parameter, bars with different letters indicate significant difference at P<0.05.
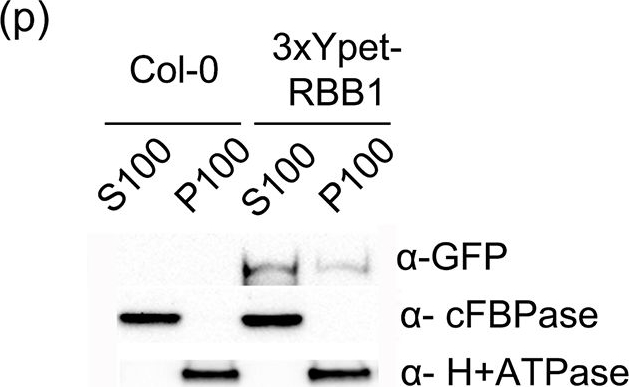
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 25915922
Journal: PLoS One
Figure Number: 6p
Published Date: 2015-04-29
First Author: Han, S. W., Alonso, J. M., et al.
Impact Factor: 2.942
Open Publication3xYpet-RBB1 is a cytoplasmic protein that associates with the tonoplast.(a-c) A 3xYpet-RBB1 fusion complements the bulb phenotype of rbb1-2. Four-day-old seedlings from rbb1-2 (a) or rbb1-2 3xYpet-RBB1 (b, c) were stained with Lysotracker Red (magenta, a, b) to label the vacuole. The signal from 3xYpet-RBB1 (c) for the complemented plant is shown. (d-f) 3xYpet-RBB1 does not co-localize with FM4-64. Images show the cotyledon of 7-d-old seedlings expressing 3xYpet-RBB1 (d) stained with 5 ?M FM4-64 (e) and the merged image (f). (g-i) 3xYpet-RBB1 and RFP-SYP22 do not co-localize. Cotyledons of 7-d-old seedlings expressing 3xYpet-RBB1 (g) and RFP-SYP22 (h) were imaged. Merged image (i) is shown. (j-l) 3xYpet-RBB1 and Lysotracker Red do not co-localize in rosette leaves. Seedlings from the 3xYpet-RBB1 line were stained with Lysotracker Red. Signal from 3xYpet-RBB1 (j), Lysotracker Red (k) and the merged image (l) are shown. Arrowheads indicate the localization of 3xYpet-RBB1 at the tip of elongating trans-vacuolar strands. (m-o) GFP molecules can also label the tips of transvacuolar strands. Seedlings from a 35::GFP marker line were stained with Lysotracker Red. The GFP signal (m), Lysotracker Red (n) and the merged image (o) are shown. Arrowheads indicate the tip of TVS. All scale bars = 20 ?m. (p) 3xYpet-RBB1 accumulates in the soluble (S100) and membrane pellet (P100) fractions from whole seedlings. Immunoblot of 3xYpet-RBB1 soluble (S100) and membrane (P100) fractions from Col-0 and 3xYpet-RBB1 transgenic plants using antibodies against GFP (?-GFP), cFBPase (?-cFBPase, control for soluble fraction), and Plasma Membrane H+ATPase (?-H+ATPase, control for membrane fraction).

Reactant: Hordeum vulgare (Barley)
Application: Western Blotting
Pudmed ID: 26213372
Journal: Sci Rep
Figure Number: 6A
Published Date: 2015-07-27
First Author: Chen, J., Wang, W. H., et al.
Impact Factor: 4.13
Open PublicationWestern-blot analysis of root PM H+-ATPase(a) and vacuolar Na+/H+ antiporter (NHE1) (c) of barley seedlings treated with 150?mM NaCl and various NaHS concentrations (0, 50, 100, 200 and 500??M) for 48?h. CK: Hoagland solution only without adding NaHS and NaCl. Three independent experiments were conducted, and the immunoblotting results show similar trends of protein expression. The relative protein expression level is shown as the ratio of PM H+-ATPase to ?-actin (b) and NHE1 to ?-actin (d) analyzed with Quantity One software. Mean values?±?SE were calculated from independent experiments and columns labeled with different letters are significantly different at P?
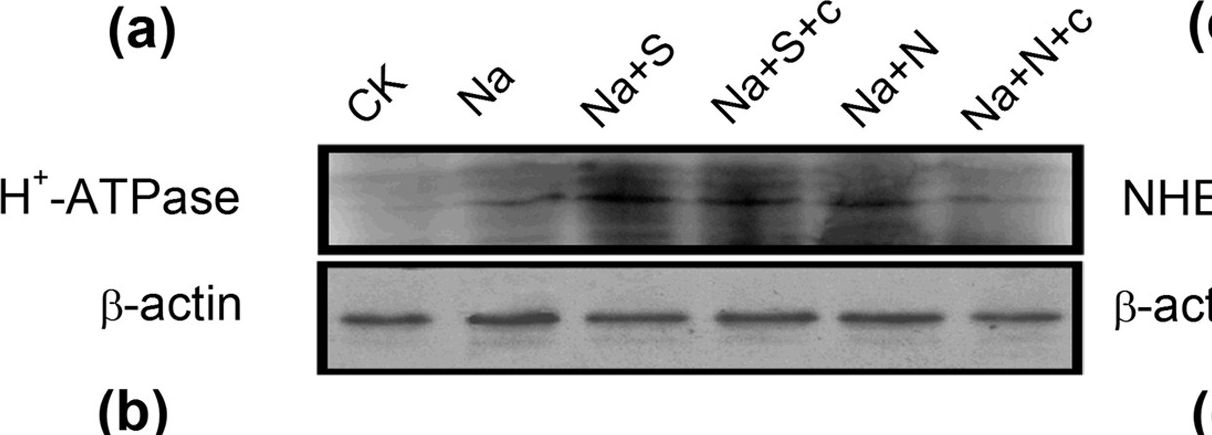
Reactant: Hordeum vulgare (Barley)
Application: Western Blotting
Pudmed ID: 26213372
Journal: Sci Rep
Figure Number: 7A
Published Date: 2015-07-27
First Author: Chen, J., Wang, W. H., et al.
Impact Factor: 4.13
Open PublicationWestern-blot analysis of root PM H+-ATPase(a) and vacuolar Na+/H+ antiporter (NHE1) (c) of barley seedlings treated with 150?mM NaCl (Na), 150?mM NaCl+100??M NaHS (Na+S), 150?mM NaCl+100??M NaHS+200??M cPTIO (Na+S+c), 150?mM NaCl+100??M SNP (Na+N), 150?mM NaCl+100??M SNP+200??M cPTIO (Na+N+c) for 48?h, respectively. CK: Hoagland solution only without adding NaHS and NaCl. Three independent experiments were conducted, and the immunoblotting results show similar trends of protein expression. The relative protein expression level is shown as the ratio of PM H+-ATPase to ?-actin (b) and NHE1 to ?-actin (d) analyzed with Quantity One software. Mean values?±?SE were calculated from independent experiments and columns labeled with different letters are significantly different at P?
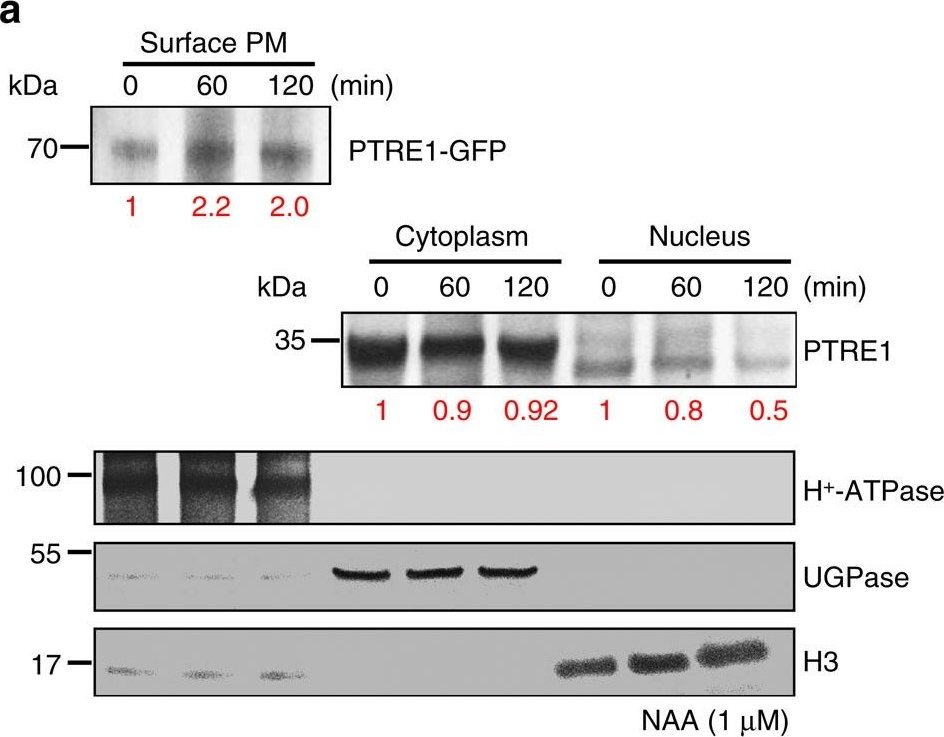
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 27109828
Journal: Nat Commun
Figure Number: 6A
Published Date: 2016-04-25
First Author: Yang, B. J., Han, X. X., et al.
Impact Factor: 13.783
Open PublicationAuxin stimulates PTRE1 accumulation at the plasma membrane.(a) Western blot analysis showed that auxin treatment results in relatively more PTRE1 at the plasma membrane, but decreased levels in nucleus and cytoplasm. Arabidopsis seedlings expressing PTRE1-GFP were treated with NAA (1??M, 60 or 120?min) and then surface-exposed membrane proteins were analysed using PTRE1 antibody or plasma membrane marker H+-ATPase. Nucleus and cytoplasm fractions were prepared from Col seedlings and analysed by western blot using antibody against PTRE1, nuclear marker H3 or cytoplasm marker UGPase. The relative quantities of the proteins were calculated by using image pro plus and indicated. (b) A proposed model for how PTRE1 and TIR1 coordinate auxin responses regulating proteasome activity and Aux/IAA protein degradation. Under normal condition (with basal auxin levels), 26S proteasome activity is maintained by appropriate distribution of PTRE1 at the plasma membrane and in intracellular compartments. In response to auxin, auxin rapidly stimulates the association of TIR1 and Aux/IAA proteins resulting in degradation of Aux/IAAs (1). Later, auxin suppresses PTRE1 to inhibit proteasome activity (possibly through stimulating the accumulation of PTRE1 at plasma membrane, resulting in decreased intracellular and nuclear localization) and hence suppresses Aux/IAA protein degradation (2) to coordinate regulation of auxin responses, reflecting a mechanism for fine control of Aux/IAA homeostasis and auxin signalling.
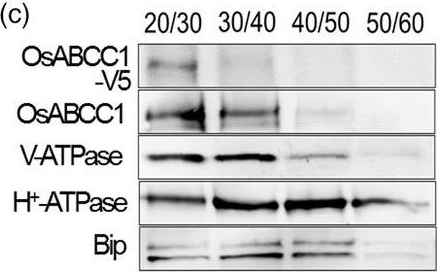
Reactant: Oryza sativa (Asian rice)
Application: Western Blotting
Pudmed ID: 29479780
Journal: Plant Biotechnol J
Figure Number: 1C
Published Date: 2018-10-01
First Author: Deng, F., Yamaji, N., et al.
Impact Factor: 8.66
Open PublicationDevelopment of transgenic rice with reduced As accumulation in the grains using tissue?specific expression of OsABCC1, ??ECS and ScYCF1. (a) Analysis of the tissue?specific expression of the OsRCc3 promoter by immunostaining OsRCc3 promoter::GUS transgenic plants (RCc3p::GUS) using a GUS antibody. The internode section denoted by broken lines is magnified in the inset surrounded by yellow solid lines. Bar = 100 ?m. (b) Maps of vectors carrying RCc3p?A (RCc3 pro::OsABCC1?V5), RCc3p?AE (RCc3 pro::OsABCC1?V5, ZmUBI pro::??ECS) or RCc3p?AEY (RCc3 pro::OsABCC1?V5, ZmUBI pro::??ECS, RCc3 pro::ScYCF1). (c) Subcellular localization of OsABCC1?V5 in the roots of RCc3p?AEY plants using a sucrose density gradient analysis. Anti?V5, anti?OsABCC1, anti?V?ATPase (tonoplast marker), anti?H+?ATPase (plasma membrane marker) and Bip (ER marker) were used to assess protein abundance in the different compartments. The signal of OsABCC1?V5 corresponded to the signals of OsABCC1 and V?ATPase. (d) Arsenic accumulation in brown rice harvested from T3 transgenic plants transformed with RCc3p?A, RCc3p?AE, RCc3p?AEY, UBIp?A, UBIp?AE or UBIp?AEY. The As concentration was analysed in brown rice from plants grown in soil with a typical basal level of As. The values are means and standard errors (n = 5 plants). Different letters indicate significantly different means (Tukey's multiple comparison analysis, P ? 0.05).
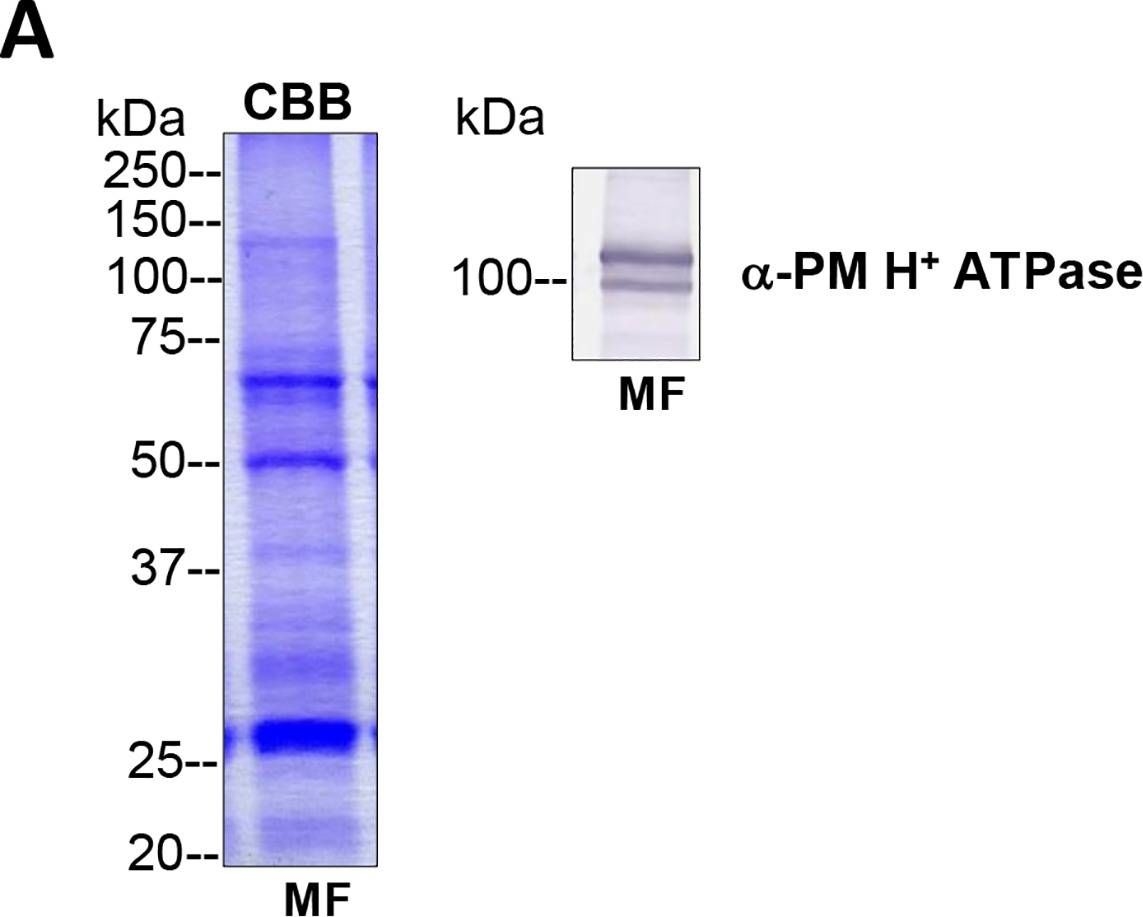
Reactant: Algae
Application: Western Blotting
Pudmed ID: 30157181
Journal: PLoS One
Figure Number: 2A
Published Date: 2018-08-30
First Author: Pertl-Obermeyer, H., Lackner, P., et al.
Impact Factor: 2.942
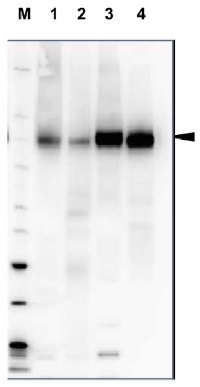
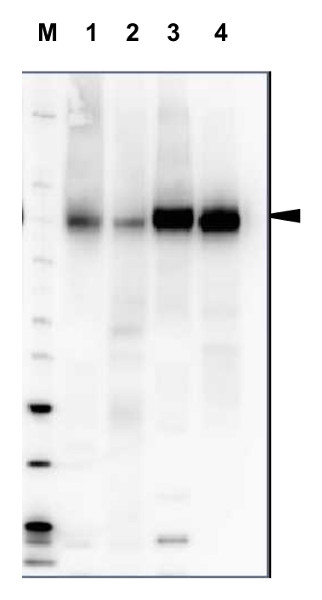
Open PublicationTotal membrane fractions of Chara internodal cells.(A) Proteins of membrane fractions (MF) were separated by SDS-PAGE (10%), stained with Coomassie Brilliant Blue (CBB) or blotted onto PVDF membranes for immunodetection of the plasma membrane H+ ATPase. 30 ?g protein per lane. Numbers on the left refer to molecular weight markers in kDa. Only the upper part of the PVDF membrane was used, the lower part was probed for immunodetection of low molecular weight proteins. (B) Immunodetection of selected organelle marker proteins for vacuoles (VHA-?, H+ PPase), ER (BiP2), plasma membrane and endosomal compartments (ARA6) or cytosol (tubulin, GRF 14-3-3). Proteins of the MFs were separated by preparative SDS-PAGE (12.75% or 10% for GRF and ARA6), plotted onto PVDF membranes cut into 3 mm strips and detected with the respective antibodies. 10 ?g protein per strip. Molecular weight markers are given in kDa. Arrow heads indicate the expected position of the respective protein.

Reactant: Algae
Application: Western Blotting
Pudmed ID: 30157181
Journal: PLoS One
Figure Number: 5B
Published Date: 2018-08-30
First Author: Pertl-Obermeyer, H., Lackner, P., et al.
Impact Factor: 2.942
Open PublicationDifferences in charasome abundance and protein expression in alkaline and acidic regions of Chara cells.(A) Left image pair: FM1-43-labelled charasomes (green fluorescence) and chloroplasts (bright field image) at an acidic band. Right image pair: FM1-43-stained charasomes are absent from the alkaline band; the bright field images show the chloroplasts. An FM1-43-stained internodal cell was cut into acid and alkaline regions as described in Materials and Methods guided by pH banding pattern visualized by phenol red. Cell fragments were mounted in artificial fresh water and examined in the CLSM. Bar = 20 ?m (B) Proteins of membrane fractions (MF) and cytosolic fractions (CF) obtained from acidic (ac) and alkaline (alk) regions were separated by SDS-PAGE (10%) and stained with Comassie Brilliant Blue (CBB) or blotted onto PVDF membranes for immunodetection of the plasma membrane H+ ATPase using only the upper part of the membrane. 15 ?g protein was loaded per lane. Numbers on the left refer to molecular weight markers in kDa. (C) Immunodetection of selected higher and lower molecular weight proteins from the membrane fraction (MF) as indicated. 10 ?g protein was loaded per lane, 10% gel. PVDF membrane was cut into upper and lower halves at 50 kDa. Molecular weight markers given in kDa.
Reactant: Vitis vinifera (Common grape vine)
Application: Western Blotting
Pudmed ID: 31156689
Journal: Front Plant Sci
Figure Number: 2E
Published Date: 2019-06-04
First Author: Kuang, L., Chen, S., et al.
Impact Factor: 5.435
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 32308664
Journal: Front Plant Sci
Figure Number: 2A
Published Date: 2020-04-21
First Author: Li, X., Sanagi, M., et al.
Impact Factor: 5.435
Akpinar, Cansev (2022) Physiological and molecular responses of roots differ from those of leaves in spinach plants subjected to short-term drought stress, South African Journal of Botany, Volume 151, Part A,2022,Pages 9-17, ISSN 0254-6299,https://doi.org/10.1016/j.sajb.2022.09.032.
Baena et al. (2022) SNARE SYP132 mediates divergent traffic of plasma membrane H+-ATPase AHA1 and antimicrobial PR1 during bacterial pathogenesis. Plant Physiol. 2022 Jun 27;189(3):1639-1661. doi: 10.1093/plphys/kiac149. PMID: 35348763; PMCID: PMC9237740.
Kostic et al. (2022),The Relative Sensitivity of Marigold vs. Tomato to Iron (Fe) Toxicity Is Associated with Root Traits: Root-to-Shoot Mass Ratio, Failure to Sequester Fe in Roots, and Levels of the Major Fe-Uptake Protein, IRT, Horticulturae 2022, 8(9), 803; https://doi.org/10.3390/horticulturae8090803
Michalopoulou et al. (2022) The host exocyst complex is targeted by a conserved bacterial type-III effector that promotes virulence. Plant Cell. 2022 May 30:koac162. doi: 10.1093/plcell/koac162. Epub ahead of print. PMID: 35640532.
Hofmann, Wienkoop & Luthje (2022) Hypoxia-Induced Aquaporins and Regulation of Redox Homeostasis by a Trans-Plasma Membrane Electron Transport System in Maize Roots. Antioxidants (Basel). 2022 Apr 25;11(5):836. doi: 10.3390/antiox11050836. PMID: 35624700; PMCID: PMC9137787.
Huang et al (2021). Parasitic modulation of host development by ubiquitin-independent protein degradation. Cell. 2021 Sep 30;184(20):5201-5214.e12. doi: 10.1016/j.cell.2021.08.029. Epub 2021 Sep 17. PMID: 34536345; PMCID: PMC8525514.
Lapshin et al. (2021) Sterol Extraction from Isolated Plant Plasma Membrane Vesicles Affects H+-ATPase Activity and H+-Transport. Biomolecules. 2021 Dec 16;11(12):1891. doi: 10.3390/biom11121891. PMID: 34944535; PMCID: PMC8699270.
Choi et al. (2021) Augmented CO2 tolerance by expressing a single H+-pump enables microalgal valorization of industrial flue gas. Nat Commun. 2021 Oct 18;12(1):6049. doi: 10.1038/s41467-021-26325-5. PMID: 34663809; PMCID: PMC8523702.
Cano-Ramirez et al. (2021) M. Plasma Membrane Fluidity: An Environment Thermal Detector in Plants. Cells. 2021 Oct 17;10(10):2778. doi: 10.3390/cells10102778. PMID: 34685758; PMCID: PMC8535034.
Adamo et al. (2021). Nanoalgosomes: Introducing extracellular vesicles produced by microalgae. J Extracell Vesicles. 2021 Apr;10(6):e12081. doi: 10.1002/jev2.12081. Epub 2021 Apr 27. PMID: 33936568; PMCID: PMC8077145.
Ekanayake et al. (2021) A. DYNAMIN-RELATED PROTEIN DRP1A functions with DRP2B in plant growth, flg22-immune responses, and endocytosis. Plant Physiol. 2021 Feb 3:kiab024. doi: 10.1093/plphys/kiab024. Epub ahead of print. PMID: 33564884.
Wang et al. (2020). Plant NLR Immune Receptor Tm-22 Activation Requires NB-ARC Domain-Mediated Self-Association of CC Domain. PLoS Pathog. 2020 Apr 27;16(4):e1008475. doi: 10.1371/journal.ppat.1008475.
Collins et al. (2020). EPSIN1 Modulates the Plasma Membrane Abundance of FLAGELLIN SENSING2 for Effective Immune Responses . Plant Physiol. 2020 Feb 24. pii: pp.01172.2019. doi: 10.1104/pp.19.01172
Zhang et al. (2018). Maintenance of mesophyll potassium and regulation of plasma membrane H+-ATPase are associated with physiological responses of tea plants to drought and subsequent rehydration. The Crop Journal July 2018. (Camellia sinensis)
Seguel et al. (2018). PROHIBITIN 3 forms complexes with ISOCHORISMATE SYNTHASE 1 to regulate stress-induced salicylic acid biosynthesis in Arabidopsis. Plant Physiol. Jan 2018. DOI:10.1104/pp.17.00941
LaMontagne et al. (2016). Isolation of Microsomal Membrane Proteins from Arabidopsis thaliana. Curr. Protoc. Plant Biol. 1:217-234. doi: 10.1002/cppb.20020.
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extracts
Plasma membrane protein isolation protocol- Reviews:
Marina Gavilanes-Ruíz | 2021-03-25The H+ATPase (AS07260) antibody was used to evaluated plasma membrane enrichment from Arabidopsis leaves. We used 1:10000 dilution in TTBS buffer supplemented with 2% (w/v) non-fat powdered milk followed for overnight at room temperature with agitation. Detection was developed with anti-rabbit IgG secondary antibody HRP conjugated (1:20000 dilution).Amelie Spazierer | 2020-07-15We used the H+ATPase antibody for Western experiments at high dilutions (1:5000) as well as for epifluorescence microscopy at lower dilutions (1:250 and 1:50) and always got a pretty good signal. There is some background for both but since the ATPase is highly abundant it is easy to get rid of that.Peeyush Ranjan | 2017-03-07The H ATPase antibody (AS07 260) was tested on total cell lysate of Chlamydomonas reinhardtii. An increasing concentration from 5ug to 50ug of total protein was loaded in 4-20% gradient gel and transferred on PVDF. After blocking with TBST 2?t free milk, membrane was incubated with 1:5000 dilution of AS07 260 Ab in TBST buffer followed by 1:10000 dilution of secondary Ab after washing. A very specific band was observed even with lowest concentration of protein. For further blotting, I used 1:7500 dilution. Very good specificity with minimum background.Anna Bilska | 2013-07-25Application: immunolocalization at the electron microscope level (immunogold), dilution: 1:100 (PBS BSA) confirmed reactivity in maize (Zea mays).Nicole Clay | 2012-12-17Antibody worked great for plasma membrane enrichment. We used 1:1000 dilution on Arabidopsis thaliana extracts that have gone through aqueous 2-phase partitioningDudley Page | 2010-05-05A strong and specific signal when used at 1/1000 against purified plasma membranes from Chlamydomonas reinhardtii.| 2009-07-21This antibody was used to detect the H ATPase in the fern Pteris vittata. In order to get decent detection of this protein, the antibody was optimized for a dilution of 1:1000. The antibody was incubated at room temperature (22 deg) for about 8 hours before washing and secondary incubation.Accessories
AS07 213 | Clonality: Polyclonal | Host: Rabbit | Reactivity: Higher plants including A.comosus, A.thaliana, C.sativus, C. australis R.Br, C.reinhardtii, F. margarita Swingle, H.vulgare, L.esculentum, L.longiflorum, Malus x domestica Borkh. c.v. Fuji, M. truncatula, M.crystallinum, N.tabacum, N.caerulescens, O.sativa, P.hybrida cv. Mitchell, Populus sp., P.vittata, Thellungiella sp., T. aestivum, Z.mays, V. vinifera | Cellular [compartment marker] of tonoplast membrane
329 €Login / Retailer